Protein N-Glycosylation
Protein N-glycosylation in bacteria and eukaryotes
One of the most abundant post-translational modifications of proteins is N-glycosylation, where complex glycans are attached to asparagine residues of proteins secreted into the lumen of the endoplasmic reticulum (ER) or into the periplasmic space of bacteria that contain N-glycosylation machinery. The central enzyme of the pathway is oligosaccharyltransferase (OST), an integral membrane protein that transfers the glycan moiety of a lipid-linked oligosaccharide (LLO) onto the acceptor asparagine of N-x-S/T sequons. The biosynthesis of LLO requires several membrane-integral or membrane-associated glycosyltransferases that facilitate the sequential assembly of the complex glycan (high-mannose glycan in eukaryotes) on a polyprenol-pyrophosphate carrier.
In 2011, we determined the first structure of a single-subunit bacterial OST enzyme, the PglB protein from the bacterium Campylobacter lari. This breakthrough, which was part of a collaboration with Markus Aebi's group (ETH Zurich) https://micro.biol.ethz.ch/research/aebi.html, enabled us to launch a comprehensive investigation into the mechanism of the OST-catalyzed reaction using biochemical, synthetic, biophysical, and structural techniques. To investigate OST function in vitro and for structural studies, we use synthetic analogs of peptides and lipid-linked oligosaccharides, which are generated by Jean-Louis Reymond's group (University of Bern) external pagehttp://www.gdb.unibe.ch/call_made. This has allowed us to trap PglB in a ternary complex with both peptide and LLO analogs bound, which revealed conformational changes in the external loop EL5 associated with substrate binding. Link to article: external pagehttps://www.nature.com/articles/nsmb.3491call_made
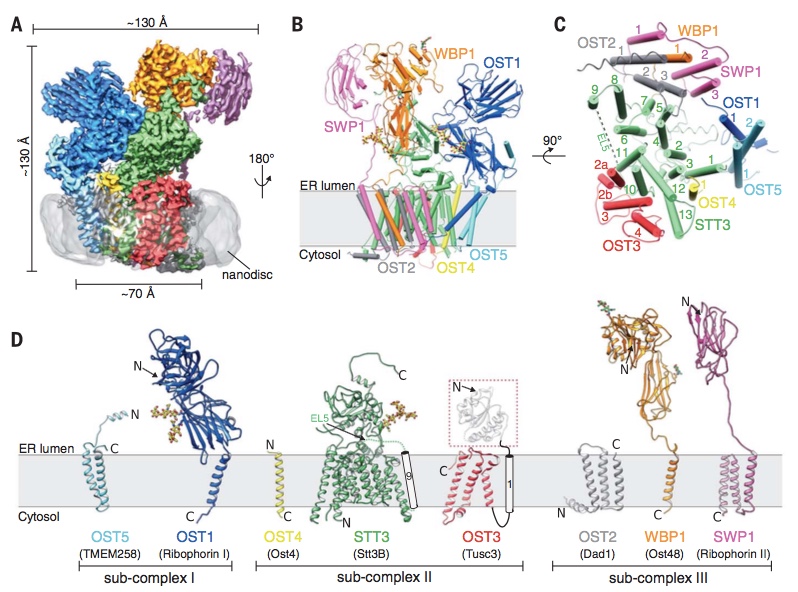
Using single particle cryo-EM, we determined the structure of the yeast OST complex to 3.3 Å resolution (Fig. 1). This structure was the first of an eukaryotic OST complex, and provided seminal insight into the architecture of eukaryotic OST and the potential functions of the non-catalytic subunits. The structure also revealed a well-ordered glycan that may have a role in substrate (LLO) binding. Link to article: external pagehttps://science.sciencemag.org/content/359/6375/545call_made
Fig. 1: Cryo-EM structure of yeast OST complex in lipidic nanodiscs (from Wild, Kowal, Eyring et al., Science 2018).
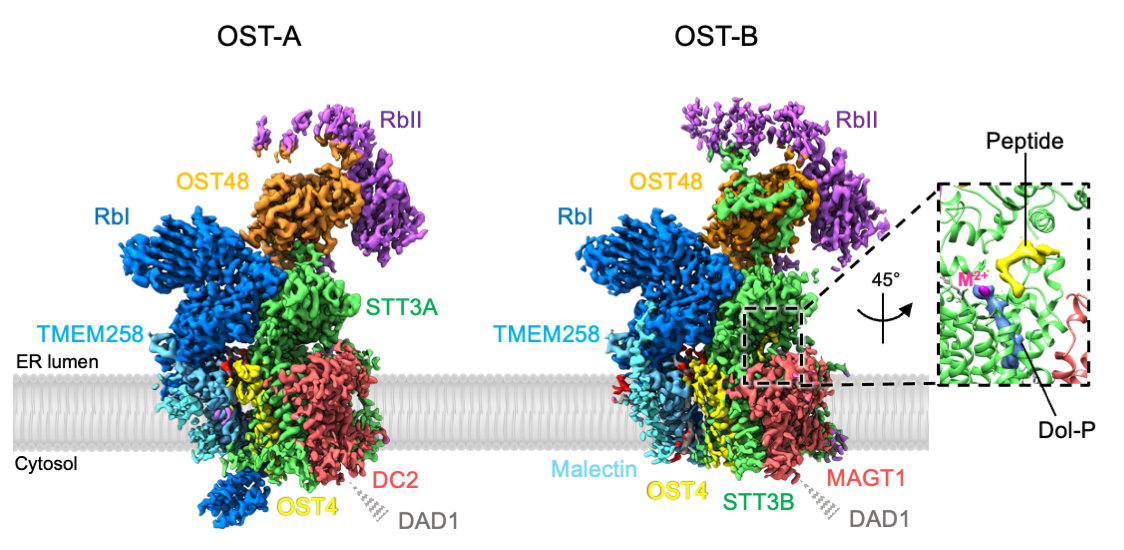
In mammals, two OST complexes coexist, which catalyze co-translational and post-translational N-glycosylation. Recently, we determined the structure of the human OST complexes OST-A and OST-B using single-particle cryo-EM (Fig. 2). Our structures revealed the molecular basis of their distinct function. Furthermore, the presence of bound ligands in the active site of OST-B gave insight into substrate binding and catalysis. Link to article: external pagehttps://science.sciencemag.org/content/366/6471/1372call_made
Fig. 2: Cryo-EM density maps of human OST-A and OST-B complexes (from Ramírez, Kowal and Locher, Science 2019).
An essential component of the protein N-glycosylation pathway is the biosynthesis of the lipid-linked oligosaccharide (LLO) that serves as the glycan donor in the OST-catalyzed reaction. A series of membrane-integral or membrane-associated glycosyltransferases (ALG proteins in eukaryotes) sequentially append saccharides to the growing glycan chain of the LLOs. A common mechanistic feature of these enzymes is the precise control of how many saccharides are added to which acceptor site on the growing glycan chain. In the bacterium Campylobacter jejuni, the membrane-associated PglH protein adds precisely 3 GalNAc moieties to the growing LLO glycan. In 2018, we established the basis of this counting mechanism by combining X-ray structures with synthetic chemistry, functional assays, and molecular dynamics simulations (collaboration with Gerhard Hummer, MPI Frankfurt) external pagehttps://www.biophys.mpg.de/en/hummer.htmlcall_made. Link to article: external pagehttps://www.nature.com/articles/s41467-018-02880-2call_made
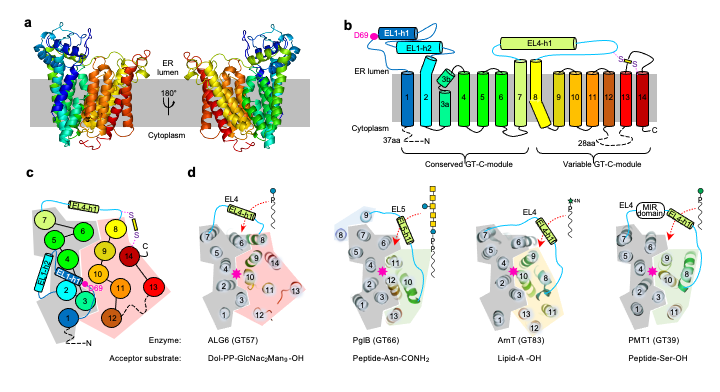
ALG6 serves in the final steps of eukaryotic LLO-biosynthesis, by adding the first of three glucoses to the emerging glycan-tree. Using synthetic substrate molecules, generated by the group of Jean-Louis Reymond external pagehttps://www.gdb.unibe.chcall_made we were able to reconstitute the ALG pathway in vitro, which enabled us to study the function of ALG6. With the help of a synthetic Fab, that was generated in collaboration with the group of external pageTony Kossiakoffcall_made external pagehttps://kosslab.uchicago.educall_made , we recently determined single-particle cryo-EM structures of apo- and donor-substrate bound ALG6 at 3.0 Å and 3.9 Å resolution, which gave first insights into its catalytic mechanism. To our surprise, the structures revealed a modular fold, which is structurally conserved among the glycosyltransferase superfamily C and explains the families’ wide range of substrate specificities. Link to article: external pagehttps://www.nature.com/articles/s41586-020-2044-zcall_made
Fig. 3: Structure and topology of the glycosyltransferase ALG6 (from Bloch et al., Nature 2020).